Second Branches
GWAS analysis of related SNP information for second branches in 115 rice accesssions
| QTL | Chr | Start | End | Sig_SNP_number | Peak_SNP | P-value | -log10(P) | Known genes |
|---|---|---|---|---|---|---|---|---|
| qSB1-1 | chr01 | 1117394 | 1142068 | 8 | 1142068 | 3.58613E-09 | 8.45 | NA |
| qSB1-2 | chr01 | 16028328 | 16428328 | 2 | 16228328 | 2.94951E-08 | 7.53 | NA |
| qSB2 | chr02 | 28071814 | 28471814 | 1 | 28271814 | 3.92377E-08 | 7.41 | NA |
| qSB3 | chr03 | 26962219 | 28511391 | 13 | 28425365 | 1.73294E-11 | 10.76 | OsTB1(LOC_Os03g49880)(Yano 2015) |
| qSB3-2 | chr03 | 20432728 | 20832728 | 1 | 20632728 | 1.64E-08 | 7.79 | NA |
| qSB4-1 | chr04 | 26628790 | 29754299 | 79 | 28823714 | 6.8576E-12 | 11.16382785 | NA |
| qSB4-2 | chr04 | 30061831 | 32604695 | 8 | 32461566 | 3.99529E-09 | 8.40 | NA |
| qSB5 | chr05 | 28154310 | 28263887 | 4 | 28154310 | 4.12558E-10 | 9.38 | NA |
| qSB6 | chr06 | 6130020 | 9424981 | 15 | 6952544 | 1.21452E-11 | 10.92 | NA |
| qSB7-1 | chr07 | 8887782 | 8981045 | 2 | 8981045 | 1.83851E-08 | 7.74 | NA |
| qSB7-2 | chr07 | 15311850 | 17127231 | 24 | 16178951 | 9.26172E-11 | 10.03 | NA |
| qSB8 | chr08 | 25464215 | 25864215 | 2 | 25664215 | 2.60795E-09 | 8.58 | NA |
| qSB10-1 | chr10 | 1111525 | 1954165 | 15 | 1139668 | 1.52133E-10 | 9.82 | NA |
| qSB10-2 | chr10 | 11690885 | 11917084 | 27 | 11871471 | 1.86617E-13 | 12.73 | NA |
| qSB11 | chr11 | 19008686 | 19666821 | 2 | 19666821 | 7.41384E-08 | 7.13 | NA |
| qSB11-2 | chr11 | 2827049 | 3227049 | 1 | 3027049 | 1.81E-07 | 6.74 | NA |
| qSB12-1 | chr12 | 551421 | 1211508 | 9 | 1019413 | 2.10861E-11 | 10.68 | NA |
| qSB12-2 | chr12 | 2026448 | 4732927 | 12 | 4717448 | 2.15907E-09 | 8.67 | NA |
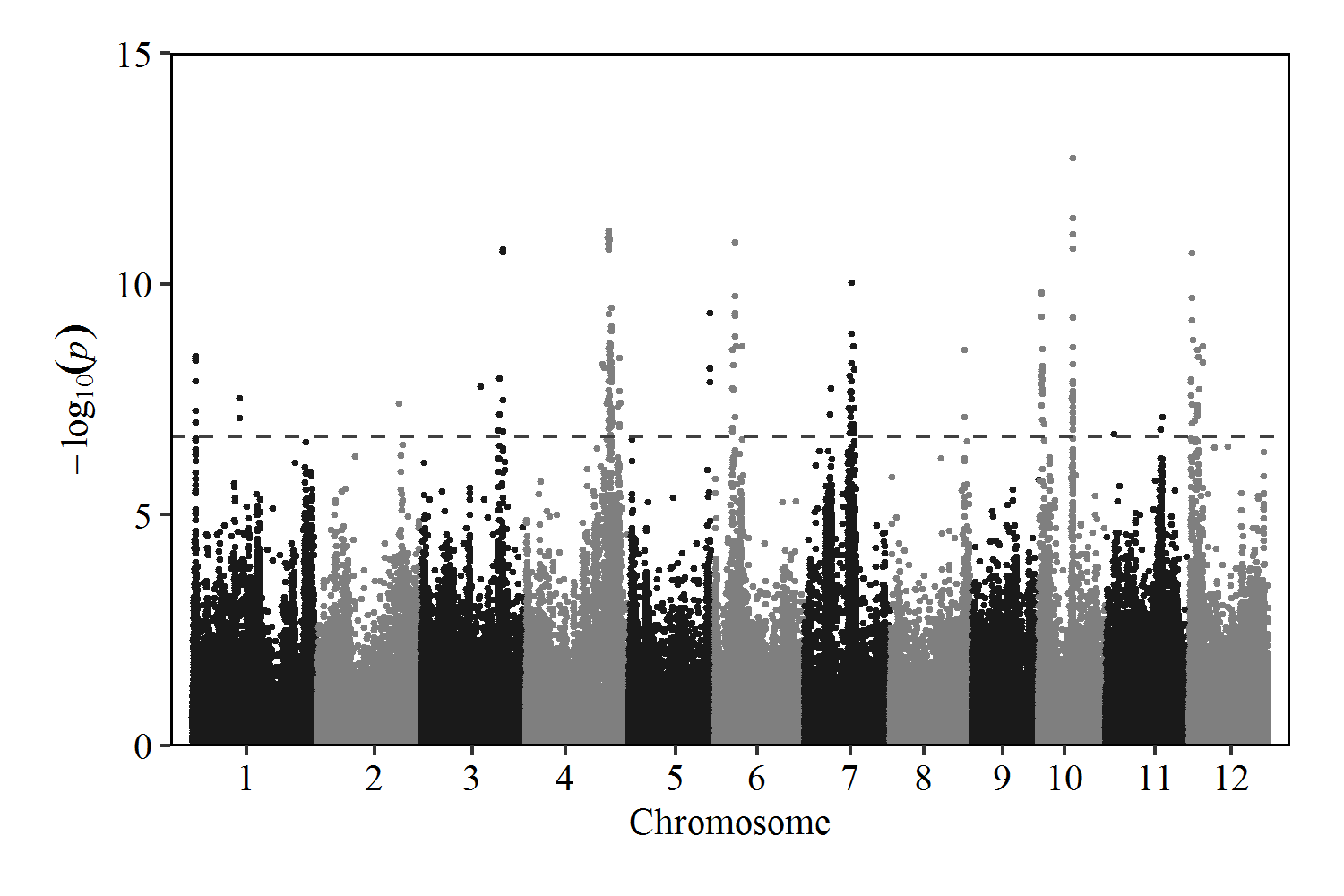
Manhattan Plot of Second Branches
QQ Plot of Second Branches
Reference
Yano K, et al. Isolation of a Novel Lodging Resistance QTL Gene Involved in Strigolactone Signaling and Its Pyramiding with a QTL Gene Involved in Another Mechanism. Mol Plant 8, 303-314 (2015).
