Panicle Length
GWAS analysis of related SNP information for panicle length in 115 rice accesssions
| QTL | Chr | Start | End | Sig_SNP_number | Peak_SNP | P-value | -log10(P) |
|---|---|---|---|---|---|---|---|
| qPL1 | chr01 | 40890615 | 41290615 | 1 | 41090615 | 1.98E-07 | 6.70 |
| qPL3 | chr03 | 14327071 | 14449173 | 3 | 14327071 | 1.7558E-08 | 7.76 |
| qPL4-1 | chr04 | 4556398 | 4956398 | 1 | 4756398 | 1.25488E-07 | 6.90 |
| qPL4-2 | chr04 | 32047542 | 32447542 | 1 | 32247542 | 8.93196E-09 | 8.05 |
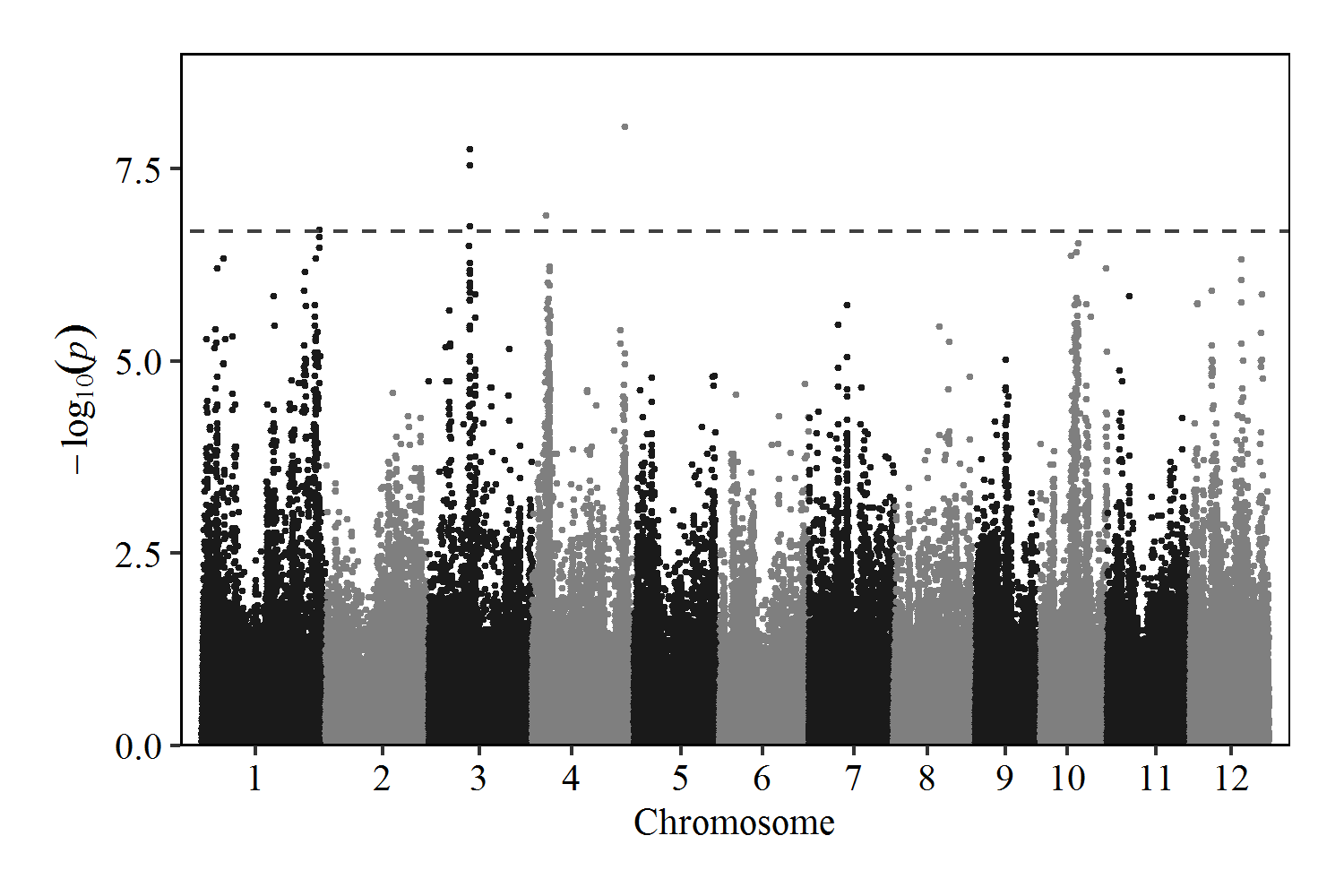
Manhattan Plot of Panicle Length
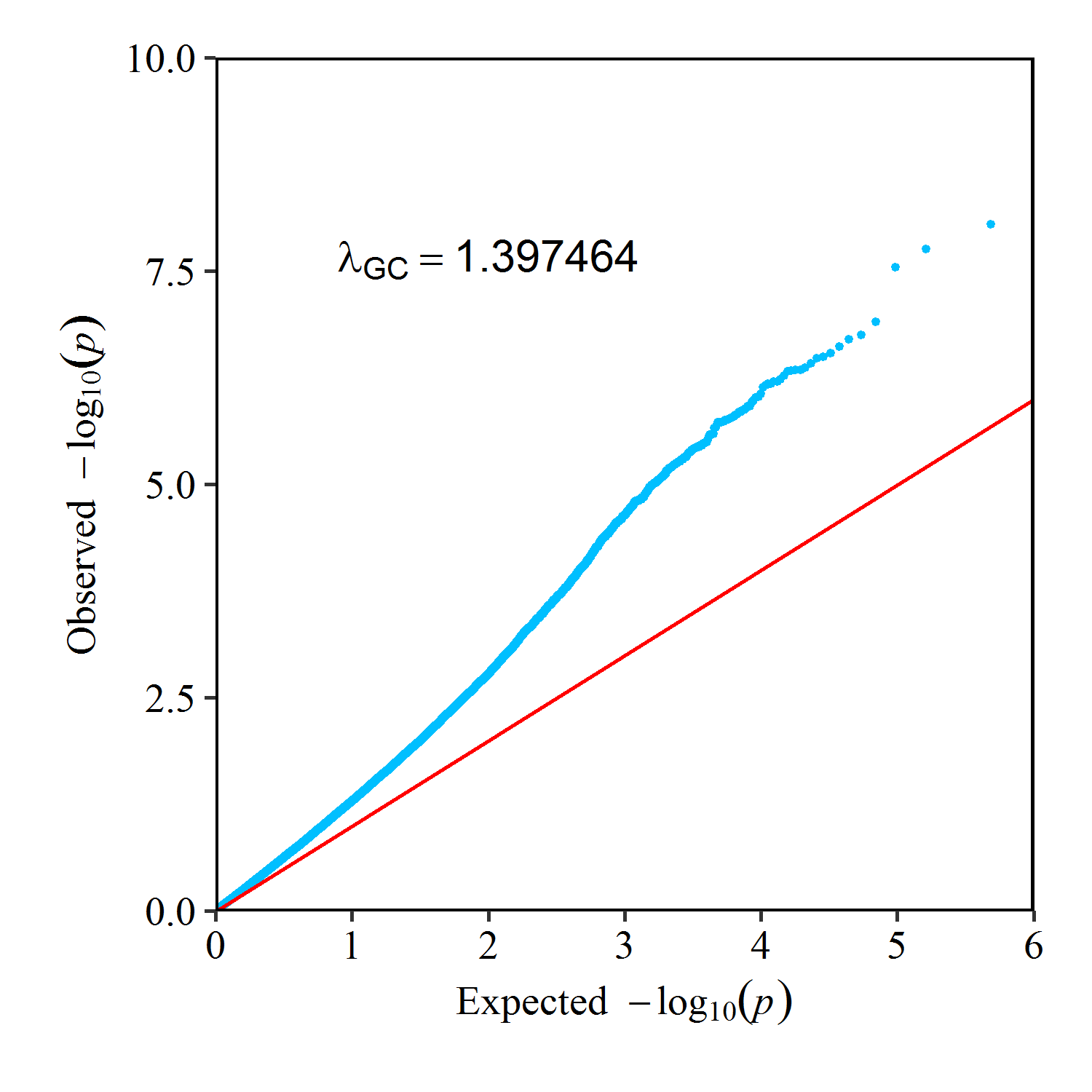
QQ Plot of Panicle Length
Panicle Length
GWAS analysis of related SNP information for panicle length in other rice accesssions
| ID | A | SE | P-Value | D | SE | P-value | h^2(a) | h^2(d) |
|---|---|---|---|---|---|---|---|---|
| Chr1-5510350-5535692 | 0.2864 | 0.0461 | 5.32E-10 | --- | --- | --- | 0.0175 | --- |
| Chr1-20054706-20059582 | 0.1074 | 0.0518 | 3.81E-02 | -0.2685 | 0.0509 | 1.32E-07 | 0.0025 | 0.0077 |
| Chr1-39315616-43201828 | 0.5405 | 0.047 | 1.46E-30 | --- | --- | --- | 0.0623 | --- |
| Chr2-5554654-5650410 | -0.2087 | 0.0529 | 8.02E-05 | --- | --- | --- | 0.0093 | --- |
| Chr2-20283991-20420828 | -0.2065 | 0.0515 | 6.07E-05 | --- | --- | --- | 0.0091 | --- |
| Chr2-33403235-33440643 | 0.2954 | 0.0506 | 5.40E-09 | --- | --- | --- | 0.0186 | --- |
| Chr3-22204599-22283352 | -0.4137 | 0.0488 | 2.50E-17 | --- | --- | --- | 0.0365 | --- |
| Chr3-24552138-24611521 | --- | --- | --- | 0.2055 | 0.052 | 7.80E-05 | --- | 0.0045 |
| Chr3-27887401-27944028 | 0.204 | 0.0525 | 1.01E-04 | 0.107 | 0.0503 | 3.33E-02 | 0.0089 | 0.0012 |
| Chr4-19211611-19218909 | 0.3687 | 0.05 | 1.69E-13 | --- | --- | --- | 0.029 | --- |
| Chr5-649060-661441 | -0.1693 | 0.0509 | 8.73E-04 | -0.103 | 0.0518 | 4.69E-02 | 0.0061 | 0.0011 |
| Chr5-17594840-18118031 | 0.3231 | 0.053 | 1.07E-09 | --- | --- | --- | 0.0223 | --- |
| Chr5-23696249-23734768 | 0.2097 | 0.0531 | 7.82E-05 | --- | --- | --- | 0.0094 | --- |
| Chr5-26344560-26371220 | 0.4604 | 0.0494 | 1.32E-20 | --- | --- | --- | 0.0452 | --- |
| Chr6-4777454-4813307 | -0.3995 | 0.0535 | 8.34E-14 | -0.1329 | 0.0494 | 7.14E-03 | 0.034 | 0.0019 |
| Chr6-5688883-6740739 | 0.3367 | 0.053 | 2.20E-10 | 0.2221 | 0.0498 | 8.19E-06 | 0.0242 | 0.0053 |
| Chr6-9660853-10010146 | 0.2888 | 0.0496 | 5.90E-09 | --- | --- | --- | 0.0178 | --- |
| Chr6-16754327-16806054 | 0.2202 | 0.049 | 7.17E-06 | --- | --- | --- | 0.0103 | --- |
| Chr6-25653684-25657376 | -0.2789 | 0.0521 | 8.89E-08 | -0.1227 | 0.0506 | 1.53E-02 | 0.0166 | 0.0016 |
| Chr6-28607084-28632979 | --- | --- | --- | 0.2456 | 0.0535 | 4.43E-06 | --- | 0.0064 |
| Chr7-28631301-28663660 | 0.206 | 0.0477 | 1.58E-05 | 0.1141 | 0.0559 | 4.14E-02 | 0.009 | 0.0014 |
| Chr8-24834539-25271327 | 0.1961 | 0.0509 | 1.16E-04 | 0.1103 | 0.0518 | 3.33E-02 | 0.0082 | 0.0013 |
| Chr9-13910718-13920635 | -0.6009 | 0.0499 | 2.99E-33 | 0.1865 | 0.0528 | 4.19E-04 | 0.077 | 0.0037 |
| Chr9-19506914-19575666 | 0.5137 | 0.0527 | 2.24E-22 | -0.1908 | 0.05 | 1.37E-04 | 0.0562 | 0.0039 |
| Chr9-21670016-21710210 | -0.3782 | 0.0517 | 2.65E-13 | 0.1722 | 0.051 | 7.31E-04 | 0.0305 | 0.0032 |
| Chr10-13030930-13067386 | -0.2711 | 0.0525 | 2.46E-07 | --- | --- | --- | 0.0157 | --- |
| Chr10-18391653-18507700 | -0.3857 | 0.0485 | 1.82E-15 | --- | --- | --- | 0.0317 | --- |
